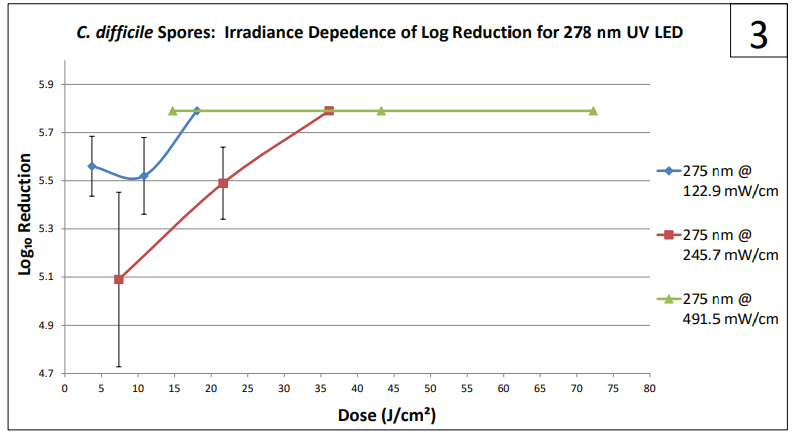
Certain clinically relevant organisms, such as Clostridium difficile (Clostridioides difficile) and Aspergillus brasilliensis, form spores that are resistant to UV inactivation. Indeed, Log₁₀ reduction in response to UV exposure is dependent not only on dose, but also on irradiance and wavelength(s) of the light. A similar observation was made when testing methods for RNase A inactivation. RNase A is an exceptionally stable enzyme that can retain much of its activity even after autoclave treatment. We found that the enzyme inactivation could be controlled by irradiance, wavelength, and dose delivered to an enzyme contaminated surface. Experiments designed to achieve high Log₁₀ reduction in CFU of spore forming microbes are time consuming, iterative, costly, require production of large numbers of spores, and must be conducted with proper biological containment. For A. brasiliensis and C. difficile the containment levels are BSL-1 and BSL-2, respectively. In this study we investigated the use of RNase A as a surrogate for estimating the effectiveness of high-intensity UV LED in reducing colony forming units of bacterial and fungal spores on surfaces, prior to culture studies.
RNase A Activity as a Predictive Tool for Log₁₀ Reduction of Bacterial and Fungal Spores on Surfaces
Categories: UV inactivation of Clostridium difficile, Laboratory Decontamination

